1. Introduction¶
This project is for Yale Course BIS 638: Clinical DMS and Ontologies.
1.1 Background¶
Ovarian cancer is the deadliest cancer of the female reproductive organs, outpacing all other forms of gynecological cancer.
According to data from national cancer institute, more than 20,000 new cases are estimated in 2020.
About 1.4 percent of women will be diagnosed with ovarian cancer at some point during their lifetime.
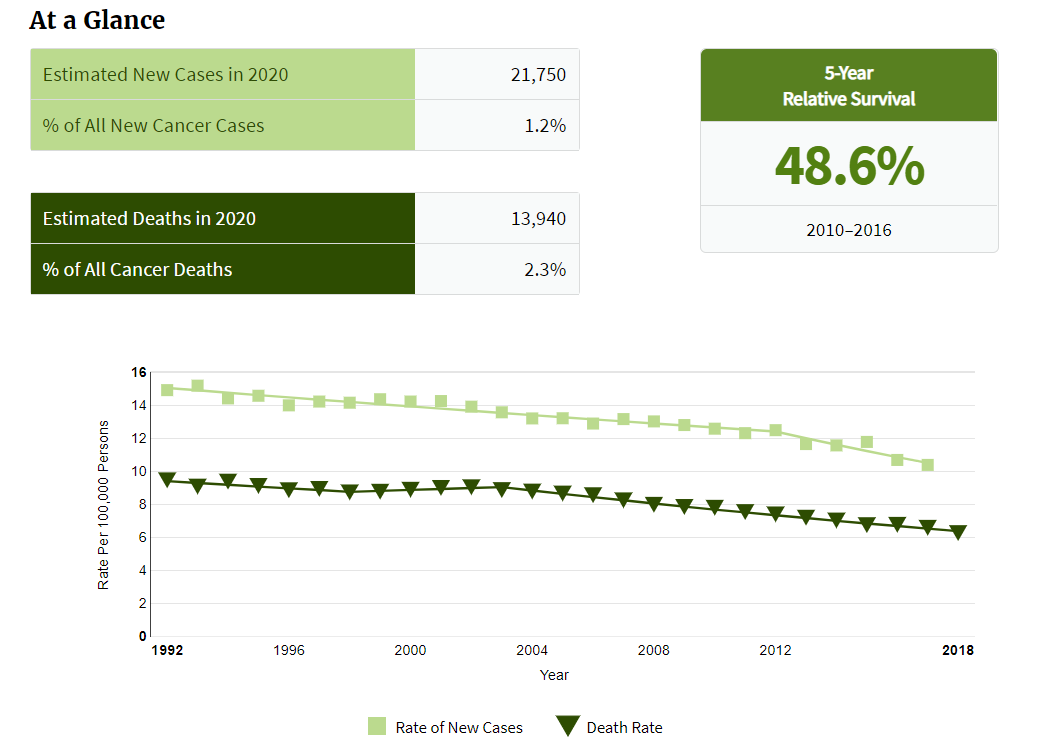 *Data source: National Cancer Institude*
*Data source: National Cancer Institude*
1.2 Research Questions¶
Do an exploratory data analysis on patients who had ovarian cancer diagnoses:
Mortality
- What’s the mortality of patients diagnosed with ovarian cancer?
- What’s the average age of death?
ICU Stay
- How long did these ovarian cancer patients stay in ICU?
- Are there any differences in the average length of stay between those who survived and died?
Standardize information using disease ontology:
- ICD9 - DOID
- Referring to previous homework, I have retrieved DOID for each icd9_code, and saved the information in
doid_icd9.csvfile.
- Referring to previous homework, I have retrieved DOID for each icd9_code, and saved the information in
- DOID - Synonym
- Referring to previous homework, I have retrieved different synonyms for each DOID, and saved the information in
synonym_full.csvfile.
- Referring to previous homework, I have retrieved different synonyms for each DOID, and saved the information in
- ICD9 - DOID
1.3 Data Sources¶
- The data comes from MIMIC-III, an openly available dataset developed by the MIT Lab for Computational Physiology, comprising deidentified health data associated with ~60,000 intensive care unit admissions. It includes demographics, vital signs, laboratory tests, medications, and more.
- To access MIMIC-III, I have finished CITI Training for Data or Specimens Only Research. Data files have be downloaded and saved in google drive, and will be loaded into google colab.

1.4 Methods¶
 |  |  |  |
- Use Vertabelo Database Modeler to design an ER model, then generate a MySQL script.
- Use google colab to install MySQL, read in SQL script and create a database with MIMIC III data.
- Use OLS as an ontology resource, link the disease terms in Disease Ontology to the patient data representation (icd9_code).
!apt install mysql-server libmysqlclient-dev
!service mysql start
!mysql -e "ALTER USER 'root'@'localhost' IDENTIFIED WITH mysql_native_password BY 'bis638'"
!pip install mysql-connector-python
- Mount google drive
from google.colab import drive
drive.mount('/content/gdrive')
- Require libraries
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from scipy import stats
2.2 Data Resource¶
- The data files we will use include:
PATIENTS: Defines each SUBJECT_ID in the database, i.e. defines a single patientADMISSIONS: Define a patient’s hospital admission, HADM_IDDIAGNOSES_ICD: Contains ICD diagnoses for patients, most notably ICD-9 diagnosesD_ICD_DIAGNOSES: Definition table for ICD diagnosesICUSTAYS: Defines each ICUSTAY_ID in the database, i.e. defines a single ICU stayPRESCRIPTIONS: Contains medication related order entries, i.e. prescriptions
- Create an ER model with Vertabelo:
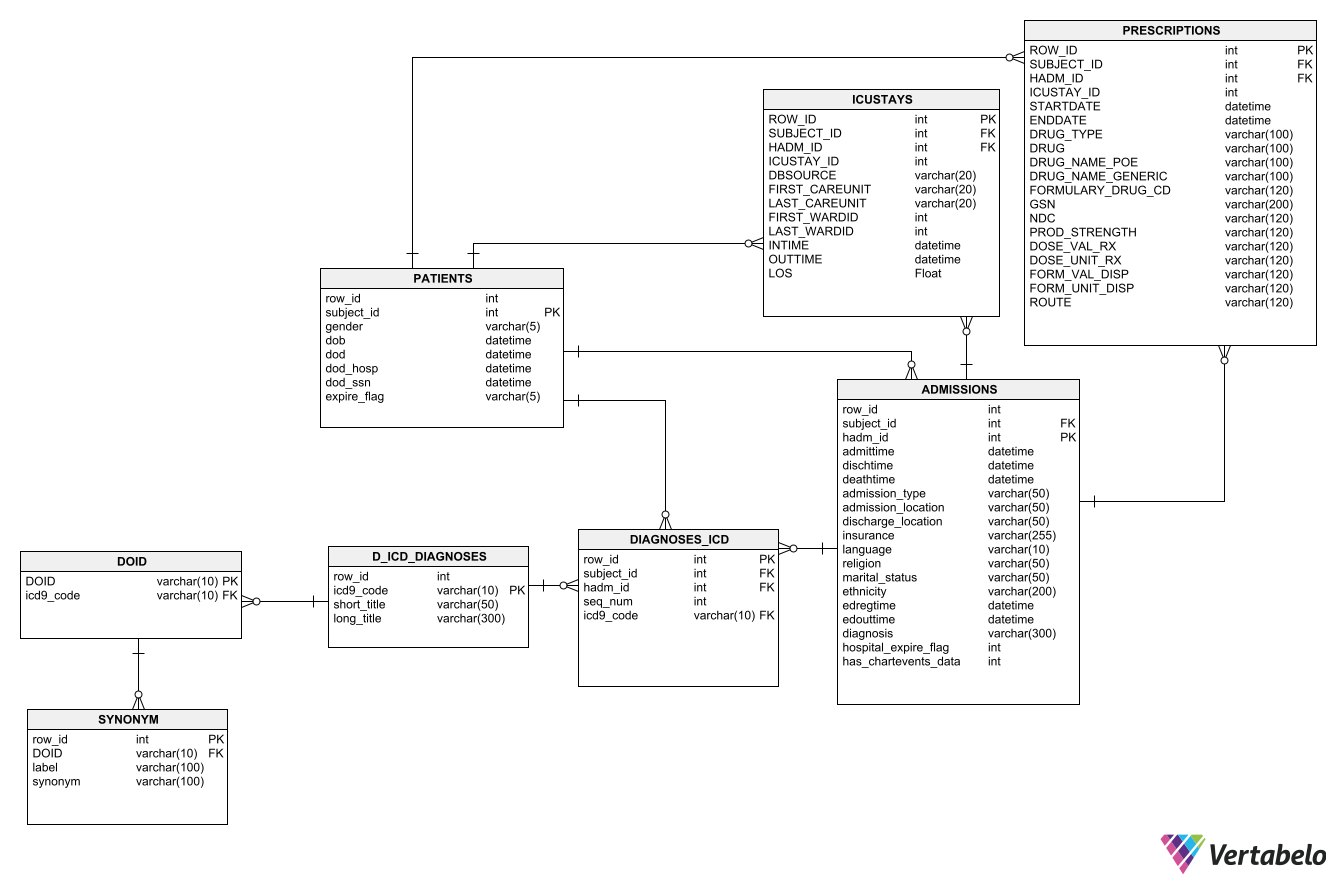
- Use the
.sqlfile generated from Vertabelo to construct the database:
!mysql -u root -p -e "CREATE DATABASE IF NOT EXISTS mimic3;\
USE mimic3;\
SOURCE /content/gdrive/My Drive/mimic_iii/MIMIC3_Project_Physical_Model_create.sql;\
COMMIT;"
- Read in the
.csvfiles into our database:
import mysql.connector
# establish a database connection with the load data file option set to True
cnx = mysql.connector.connect(user='root',
password='bis638',
host='127.0.0.1',
database='mimic3',
allow_local_infile=True)
# create database_cursor to perform SQL operation
db_cursor = cnx.cursor()
# execute the sql command to load a data file into a table (repeat the following line for more data files and tables)
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/PATIENTS.csv' into table PATIENTS fields terminated by ',' ignore 1 lines")
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/ADMISSIONS.csv' into table ADMISSIONS fields terminated by ',' ignore 1 lines")
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/ICUSTAYS.csv' into table ICUSTAYS fields terminated by ',' ignore 1 lines")
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/PRESCRIPTIONS.csv' into table PRESCRIPTIONS fields terminated by ',' ignore 1 lines")
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/D_ICD_DIAGNOSES.csv' into table D_ICD_DIAGNOSES fields terminated by ',' ignore 1 lines")
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/DIAGNOSES_ICD.csv' into table DIAGNOSES_ICD fields terminated by ',' ignore 1 lines")
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/DOID.csv' into table DOID fields terminated by ',' ignore 1 lines")
db_cursor.execute("Load data local infile '/content/gdrive/My Drive/mimic_iii/synonym_full.csv' into table SYNONYM fields terminated by ',' ignore 1 lines")
# commit the loading data transaction
db_cursor.execute("commit")
2.3 Ovarian Cancer Patients¶
- We want to extract information that only contains ovarian cancer patients.
- First, we need to know what
icd9_coderepresents ovarian cancer.
- First, we need to know what
!mysql -u root -p -e "USE mimic3;\
SELECT * FROM D_ICD_DIAGNOSES \
WHERE (long_title LIKE '%OVARY%' OR long_title LIKE '%OVARIAN%') \
AND (long_title LIKE '%CANCER%' OR long_title LIKE '%NEOPLASM%');"
After manully screening, we decide to include patients with
icd9_codeof1830,1986, andV1043as ovarian cancer patients.Extract all the ovarian cancer patients (but only show the first 20 records and selected parameters).
!mysql -u root -p -e "USE mimic3;\
CREATE TABLE IF NOT EXISTS ovarian_cancer AS \
(SELECT a.subject_id, a.hadm_id, a.diagnosis, a.marital_status, a.ethnicity, a.hospital_expire_flag, di.icd9_code FROM ADMISSIONS AS a \
INNER JOIN DIAGNOSES_ICD AS di \
ON a.hadm_id = di.hadm_id \
WHERE di.icd9_code IN ('\"1830\"', '\"1986\"', '\"V1043\"') \
ORDER BY subject_id);"
!mysql -u root -p -e "USE mimic3;\
SELECT o.subject_id, o.hadm_id, o.diagnosis, o.icd9_code, p.expire_flag FROM ovarian_cancer o \
INNER JOIN PATIENTS p ON o.subject_id = p.subject_id \
LIMIT 20;"
- There are in total 169 patients in MIMIC3 database who had ovarian cancer, and they made about 200 admissions in total.
!mysql -u root -p -e "USE mimic3;\
SELECT COUNT(DISTINCT subject_id) AS ovarian_cancer_subject, COUNT(DISTINCT hadm_id) AS ovarian_cancer_admission FROM ovarian_cancer;"
sql_statement = "SELECT expire_flag FROM PATIENTS \
WHERE subject_id in (SELECT DISTINCT subject_id FROM ovarian_cancer)"
db_cursor.execute(sql_statement)
dbresult = db_cursor.fetchall()
death_indicator = []
for r in dbresult:
death_indicator.append(r[0])
# print(death_indicator)
survive = death_indicator.count('0')
death = death_indicator.count('1')
plt.pie([survive, death], labels = ['survive', 'dead'], autopct = '%1.2f%%')
plt.title('Mortality of Ovarian Cancer Patients')
plt.show()
- Then, we want to know the average age of those who died.
- Before the calculation, take a look at each patient's death age.
- Some patients has a death age of 300, which is obviously impossible. Therefore, we do not account for this patient in our following analysis.
- Only show the first 20 records here.
!mysql -u root -p -e "USE mimic3;\
SELECT subject_id, dob, dod, round(left((dod-dob), length(dod-dob)-8)/100) AS expire_age FROM PATIENTS \
WHERE subject_id in (SELECT DISTINCT subject_id FROM ovarian_cancer) \
AND expire_flag = 1 \
LIMIT 20;"
- Use a histgram to intuitively show the pattern.
sql_statement = "SELECT round(left((dod-dob), length(dod-dob)-8)/100) AS expire_age FROM PATIENTS \
WHERE subject_id in (SELECT DISTINCT subject_id FROM ovarian_cancer) \
AND expire_flag = 1 \
AND left((dod-dob), length(dod-dob)-8)/100 < 100"
db_cursor.execute(sql_statement)
dbresult = db_cursor.fetchall()
ages = []
for r in dbresult:
ages.append(r[0])
plt.hist(ages, bins = 12, color = 'steelblue', edgecolor = 'k')
plt.title('Age of death distribution for Ovarian Cancer Patients')
plt.show()
- Calculate the average expire age.
!mysql -u root -p -e "USE mimic3;\
SELECT round(AVG(left((dod-dob), length(dod-dob)-8)/100)) AS avg_expire_age FROM PATIENTS \
WHERE subject_id in (SELECT DISTINCT subject_id FROM ovarian_cancer) \
AND expire_flag = 1 \
AND left((dod-dob), length(dod-dob)-8)/100 < 100;"
3.2 ICU Stay¶
- We want to know whether these patients diagnosed with ovrian cancer had ICU experiences.
- How long did they stay?
- Are there any differences in the length of stay between those who survived and died?
- First, calculate the lenth of stay for each ICU admission.
- Only show the first 20 records here.
!mysql -u root -p -e "USE mimic3;\
CREATE TABLE IF NOT EXISTS ovarian_icu AS \
(SELECT i.subject_id, i.hadm_id, i.icustay_id, i.los, p.expire_flag FROM ADMISSIONS AS a \
LEFT JOIN ICUSTAYS AS i ON i.hadm_id = a.hadm_id \
LEFT JOIN PATIENTS AS p ON p.subject_id = a.subject_id \
WHERE i.hadm_id in (SELECT hadm_id FROM ovarian_cancer)); \
SELECT * FROM ovarian_icu \
LIMIT 20;"
- Note that some patients may stay in ICU for several times.
- Therefore, we want to modify the above table, to calculate a total stay length for each subject.
- Only show the first 20 records here.
!mysql -u root -p -e "USE mimic3;\
CREATE TABLE IF NOT EXISTS ovarian_los AS \
(SELECT oi.subject_id, ROUND(SUM(oi.los), 1) AS total_los, p.expire_flag FROM ovarian_icu AS oi \
INNER JOIN PATIENTS AS p ON oi.subject_id = p.subject_id \
GROUP BY subject_id); \
SELECT * FROM ovarian_los \
LIMIT 20;"
- The average length of stay in ICU is 3.3 days for the survived and 4.7 for the died.
!mysql -u root -p -e "USE mimic3;\
SELECT expire_flag, ROUND(AVG(total_los),1) AS average_los FROM ovarian_los GROUP BY expire_flag;"
- We want to do a statistical test to see if there is a significant difference between those who survived and died.
### need to close the cursor and reconnect to avoid potential bug
db_cursor.close()
cnx.close()
import mysql.connector
cnx = mysql.connector.connect(user='root', password='bis638', host='127.0.0.1', database='mimic3', allow_local_infile=True)
db_cursor = cnx.cursor()
sql_statement = "SELECT * FROM ovarian_los"
db_cursor.execute(sql_statement)
dbresult = db_cursor.fetchall()
# initiate a temporary list to save the dictionaries
tmp = []
# initiate an index
i = 0
for r in dbresult:
LOS = {}
LOS['subject_id'] = r[0]
LOS['total_los'] = r[1]
LOS['expire_flag'] = r[2]
# save the value of dictionary in the list
tmp.append(LOS.values())
i = i + 1
df = pd.DataFrame(columns = ['subject_id', 'total_los', 'expire_flag'], data = tmp)
rvs1 = df.loc[df["expire_flag"] == "1", ["total_los"]]
rvs0 = df.loc[df["expire_flag"] == "0", ["total_los"]]
# Do the levene test to assess the equality of variances
stats.levene(rvs1['total_los'], rvs0['total_los'])
# LeveneResult p > 0.05, we can assume that variances are the same
# Do the t test
stats.ttest_ind(rvs1['total_los'], rvs0['total_los'])
- p = 0.13 > 0.05, we fail to reject the null hypothesis and conclude that there is no significant difference in average length of ICU stay between those who survived and those who died.
4. Ontology¶
- I tried to retrieve the DOID for each icd9_code, but turned out there was no corresponding DOID for secondary malignant neoplasm of ovary and personal history of ovarian neoplasm.
!mysql -u root -p -e "USE mimic3;\
CREATE TABLE IF NOT EXISTS ovarian_syn AS \
(SELECT DISTINCT did.icd9_code, did.short_title, did.long_title, DOID.DOID, SYNONYM.label, SYNONYM.synonym FROM D_ICD_DIAGNOSES AS did \
LEFT JOIN DOID ON did.icd9_code = DOID.icd9_code \
LEFT JOIN SYNONYM ON DOID.DOID = SYNONYM.DOID \
WHERE did.icd9_code LIKE '_1830_' \
OR did.icd9_code LIKE '_1986_' \
OR did.icd9_code LIKE '_V1043_'); \
SELECT * FROM ovarian_syn;"
- Here comes the problem:
- When I try to retrieve patients with malignant tumour of ovary, not only patients with
icd9_code = 1830meet the criteria; I also want to retrieve patients withicd9_code = 1986. - But when I try to retrieve patients with primary ovarian cancer, I do not want patients with
icd9_code = 1986 (Secondary malignant neoplasm of ovary).
- When I try to retrieve patients with malignant tumour of ovary, not only patients with
- Therefore, I would like to do a little modification to the above table.
!mysql -u root -p -e "USE mimic3;\
START TRANSACTION; \
INSERT INTO ovarian_syn VALUES ('\"1986\"', 'Second malig neo ovary', 'Secondary malignant neoplasm of ovary', 'NULL', 'NULL', 'malignant tumour of ovary'); \
INSERT INTO ovarian_syn VALUES ('\"1986\"', 'Second malig neo ovary', 'Secondary malignant neoplasm of ovary', 'NULL', 'NULL', 'ovarian neoplasm'); \
INSERT INTO ovarian_syn VALUES ('\"1986\"', 'Second malig neo ovary', 'Secondary malignant neoplasm of ovary', 'NULL', 'NULL', 'tumor of the Ovary'); \
INSERT INTO ovarian_syn VALUES ('\"1986\"', 'Second malig neo ovary', 'Secondary malignant neoplasm of ovary', 'NULL', 'NULL', 'malignant Ovarian tumor'); \
INSERT INTO ovarian_syn VALUES ('\"1986\"', 'Second malig neo ovary', 'Secondary malignant neoplasm of ovary', 'NULL', 'NULL', 'ovary neoplasm'); \
COMMIT; \
SELECT * FROM ovarian_syn WHERE synonym NOT IN ('NULL');"
- Finally, we can use synonyms to retrieve corresponding subjects and admissions as we expected.
- Sample code as below (the results are too long so I hide them).
!mysql -u root -p -e "USE mimic3;\
SELECT diag.subject_id, diag.hadm_id, diag.icd9_code FROM ovarian_syn s \
INNER JOIN DIAGNOSES_ICD diag on s.icd9_code = diag.icd9_code \
WHERE s.synonym = 'malignant tumour of ovary';"
5. Reference & Acknowlegement¶
Some part of inspirations comes from 44 Statistics to Know About Ovarian Cancer.
Thanks to Kei-Hoi, David, and Vimig, for this fantastic, brand-new course.